|
A biological system is robust if it continues to function in the face of perturbations. The perturbations can be genetic, i.e., mutations, or environmental, e.g. changes in temperature or pH. Under what circumstances can robustness evolve? How does it affect the potential for future innovation in evolution? These are some of the questions we explore using RNA secondary structure as a model molecular phenotype.
Traditionally population genetics considers the evolution of a limited number of genotypes corresponding to phenotypes with different fitness. As model phenotypes, in particular RNA secondary structure, have become computationally tractable, however, it has become apparent that the context dependent effect of mutations and the many-to-one nature inherent in these genotype-phenotype maps can have fundamental evolutionary consequences. In particular, it has been shown that populations of genotypes evolving on neutral networks, corresponding to genotypes with the same secondary structure, can evolve mutational robustness [Nimwegen et al. 1999, PNAS 96:9716] by concentrating the population in regions of high neutrality.
A property of fundamental interest that can be investigated using RNA secondary structure as a model molecular phenotype is genetic robustness, that is the preservation of an optimal phenotype in the face of mutations. Genetic robustness is critical to the understanding of evolution, as phenotypically expressed genetic variation is the fuel of natural selection. The origin of genetic robustness, whether it evolves directly by natural selection or it is a correlated byproduct of other phenotypic traits, is unresolved. Examining microRNA (miRNA) genes of several eukaryotic species, Borenstein and Ruppin [Borenstein and Ruppin 2006, PNAS 103: 6593] showed that the structure of miRNA precursor stem-loops exhibits significantly increased mutational robustness in comparison with a sample of random RNA sequences with the same stem-loop structure. The observed robustness was found to be uncorrelated with traditional measures of environmental robustness - implying that miRNA sequences show evidence of the direct evolution of genetic robustness. These findings are surprising, as theoretical results indicate that the direct evolution of robustness requires high mutation rates and/or large effective population sizes only found among RNA viruses, not multicellular eukaryotes.
Extending our theoretical understanding by examining the role of recombination, we have demonstrated that while mutational robustness is significantly enhanced in the presence of recombination, large effective population sizes and/or mutation rates are still required for mutational robustness to evolve directly. We further demonstrated that the sampling method used by Borenstein and Ruppin introduced significant bias that lead to an overestimation of robustness.
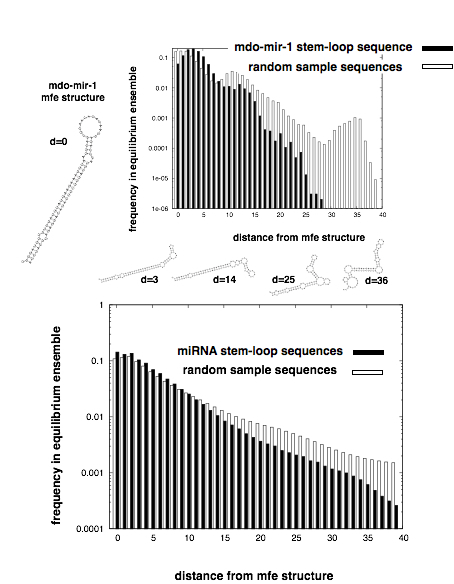
Introducing a novel measure of robustness based on the equilibrium thermodynamic ensemble of secondary structures of the miRNA precursor sequences, we were able to demonstrate that, (i) miRNA are significantly more tolerant with respect to thermal and mutational perturbations (exhibit enhanced thermodynamic and mutational robustness) than samples of random RNA sequences with the same MFE structure, (ii) the biophysics of RNA folding induces a high level of correlation between the responses to mutational and thermodynamic perturbations. Our results, based on the computational analysis of an exhaustive set of miRNA genes, strongly suggest that the successful processing of miRNA sequences is not determined by strict adherence to the MFE structure, but rather by the criterion that the RNA sequence fold into a structure with sufficient similarity to the MFE structure with high enough probability.
Publications:
Szollosi GJ, Derenyi I (2008) The Effect of Recombination on the Neutral Evolution of Genetic Robustness. Math Biosci; arxiv;
Szollosi GJ, Derenyi I (2009) Congruent evolution of genetic and environmental robustness in microRNA. Mol Biol Evol; arxiv;
Szollosi GJ, Derenyi I (in preperation) The effective temperature of mutations and the congruent evolution of robustness
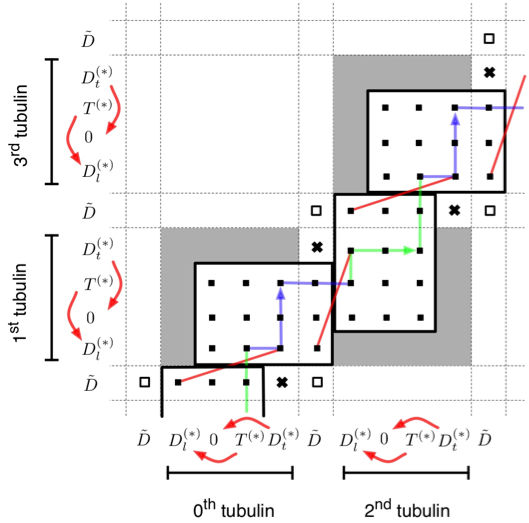
In our work on kinesin we have endeavoured to develop a thermodynamically consistent model of the kinetics of kinesin under load, using an explicit statistical physical description of the neck-linkers of the kinesin homodimer.
Conventional kinesin is a two-headed homodimeric motor protein, which is able to walk along microtubules processively by hydrolyzing ATP. Neck linker docking is widely believed to be a crucial element of this mechanism, without which the experimentally observed dwell times of the steps could not be explained. Using results from the theory of polymers we have show that the experimental data impose very strict constraints on the lengths of both the neck linker and its docking section.
The neck linkers of kinesin, which connect the two motor domains and can undergo a docking/undocking transition, have also been postulated to play the key role in the coordination of the chemical cycles of the two motor domains and, consequently, in force production and directional stepping. Although many experiments, often complemented with partial kinetic modeling of specific pathways, support this idea, the ultimate test of the viability of this hypothesis requires the construction of a complete kinetic model. We show that by considering the two neck linkers as entropic springs that are allowed to dock to their head domains, and incorporating only the few most relevant kinetic and structural properties of the individual heads, it is possilbe to assemble the first detailed, thermodynamically consistent model of dimeric kinesin that can (i) explain the cooperation of the heads during walking and (ii) reproduce much of the available experimental data (speed, dwell time distribution, randomness, processivity, hydrolysis rate, etc.) under a wide range of conditions (nucleotide concentrations, loading force, neck linker length and composition, etc.) simultaneously. Besides revealing the mechanism by which kinesin operates, our model also makes it possible to look into the experimentally inaccessible details of the mechanochemical cycle and predict how certain changes in the protein would affect its motion.
Publications:
Czovek A, Szollosi GJ, Derenyi I (2008) The relevance of neck linker docking in the motility of kinesin. Biosystems; arxiv;
Czovek A, Szollosi GJ, Derenyi I
(2011) to appear BioPhysJ
video 1; video 2; online kinetics;
Evolutionary dynamics acts, on populations, not individuals, and the realization that frequency dependent fitness introduces strategic aspects to evolution has lead to the emergence of an broadl influential interdisciplinary field -- evolutionary game theory. More recently the investigation of the evolutionary dynamics of populations with spatial structure has lead to the recognition that the success of different strategies can be fundamentally influenced by the topology of interactions. The effects of spatial structure in model systems have, however, proved to be sensitive to the details of the underlying topology and dynamics.
We introduce a minimal model of population structure that is described by two distinct hierarchical levels of interaction: population size at the local scale and the relative strength of local dynamics to global mixing. Applying our model of spatial structure to the repeated prisoner's dilemma we uncover a novel and counterintuitive mechanism by which the constant influx of defectors facilitates the survival of reciprocity through stabilizing cycles of cooperation, reciprocity and defection. Further exploring the phase space of the repeated prisoner's dilemma and also of the ``rock-paper-scissor'' games we are able to reproduce effects observed in models with explicit spatial embedding, such as the maintenance of biodiversity and the emergence of global oscillations.This minimal population structure we describe is more relevant in a wide range of natural systems, than more subtle setups with a delicate dependence on the details of the topology and can reveal universal effects of spatial structure that do not depend on these details.
Publication:
Szollosi GJ, Derenyi I (2008) Evolutionary games on minimally structured populations. Phys Rev E; arxiv;
|

![]()
 ORCiD ID:
ORCiD ID:  Google Scholar:
Google Scholar: