Szollosi GJ's Home Page
at Eötvös University
| past projects |
| cv |
| publications |
| contact |
| Hi, |
My name is Gergely Szöllősi and I currently head the "Lendület" Evolutionary Genomics Research Group of the Hungarian Academy of Sciences and the ERC "GENECLOCKS" Research Group hosted at The Institute of Physics at Eötvös University in Budapest. Until 2013 July I was a a Marie Curie Fellow at the Bioinformatics. Phylogenetics and Evolutionary Genomics Group at LBBE in Lyon. For more details about me and my career to date see my CV and here. NEWS 2024: Join the Model-Based Evolutionary Genomics Unit at Okinawa the Institute of Science and Technology for as a postdoc, senior postdoc or scientific programmer working at the intersection of computational and evolutionary biology. Our unit works to decode the complex patterns and processes that underpin the evolution of life across its myriad forms with a significant focus on reconstructing the Tree of Life and modeling the dependency between different levels of organization in biological systems. By employing probabilistic models and leveraging the power of machine learning, we aim to unravel the co-evolutionary dynamics that have shaped the Tree of Life, from the enigmatic origins of early life forms to the sophisticated structures of modern biological systems. 
Congratulations to Lénárd for winning the Publisher's Award for Excellence in Systematic Research with his paper on how compositionally constrained sites drive long-branch attraction, wherein, according to the award committee: Lenard and collaborators investigate the link between long-branch attraction and across-site compositional heterogeneity in deep phylogenetics via simulations and empirical datasets. They also propose a new pipeline based on the CAT model (CAT-PMSF) to assess the risk of compositional heterogeneity bias when the CAT model does not converge. Given the computational effort required to run a CAT analysis on a large genomic dataset, this work is particularly relevant. The authors do not only offer a tool to help researchers be aware of the effects of this model deviation on phylogenetic accuracy, but also a way to solve it. 2023: Starting from October 1st 2023 I am joining Okinawa Institute of Science and Technology as a Transitional Associate Professor leading the Model-Based Evolutionary Genomics Unit. The unit will start full time from October 1st 2024, annocment for several postdoc positions coming soon!
AleRax is out! Thanks to great work by Benoit Morel at Alexis' lab it is 10x faster 🚀 than ALE, but just as accurate. It can also infer a species tree 🌲 and, most importantly, process gene families in parallel to estimate lineage specific DTL parameters 🎄 for genome-scale datasets with 100s of taxa. 2022: Congratulations to Edu for his first publication as a postdoc! A strong start with a Nature paper: "Divergent genomic trajectories predate the origin of animals and fungi". See also the University news piece in English and Hungarian! 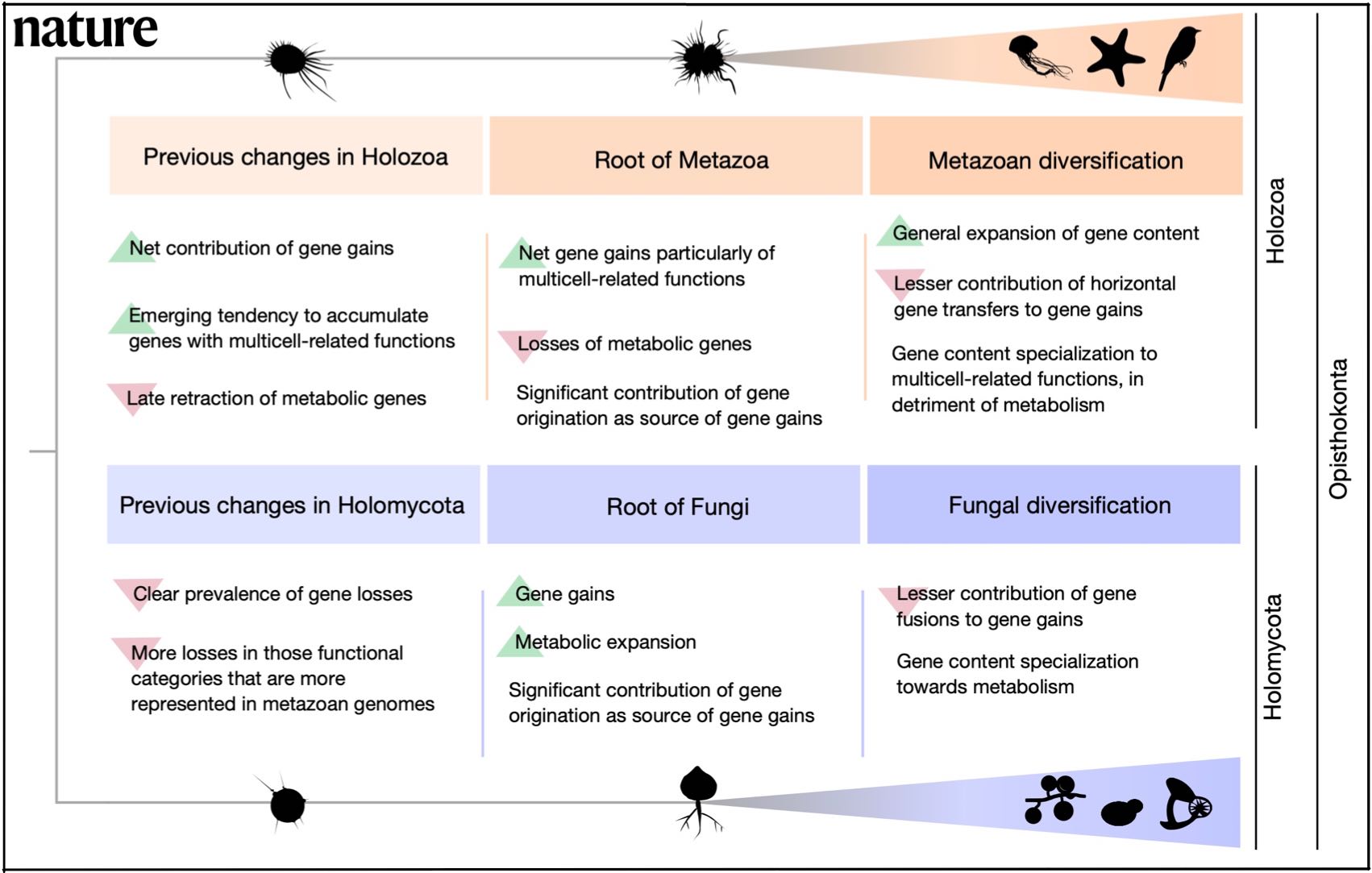
Our new paper "Trade-off between reducing mutational accumulation and increasing commitment to differentiation determines tissue organization" is out in Nature Communcations! 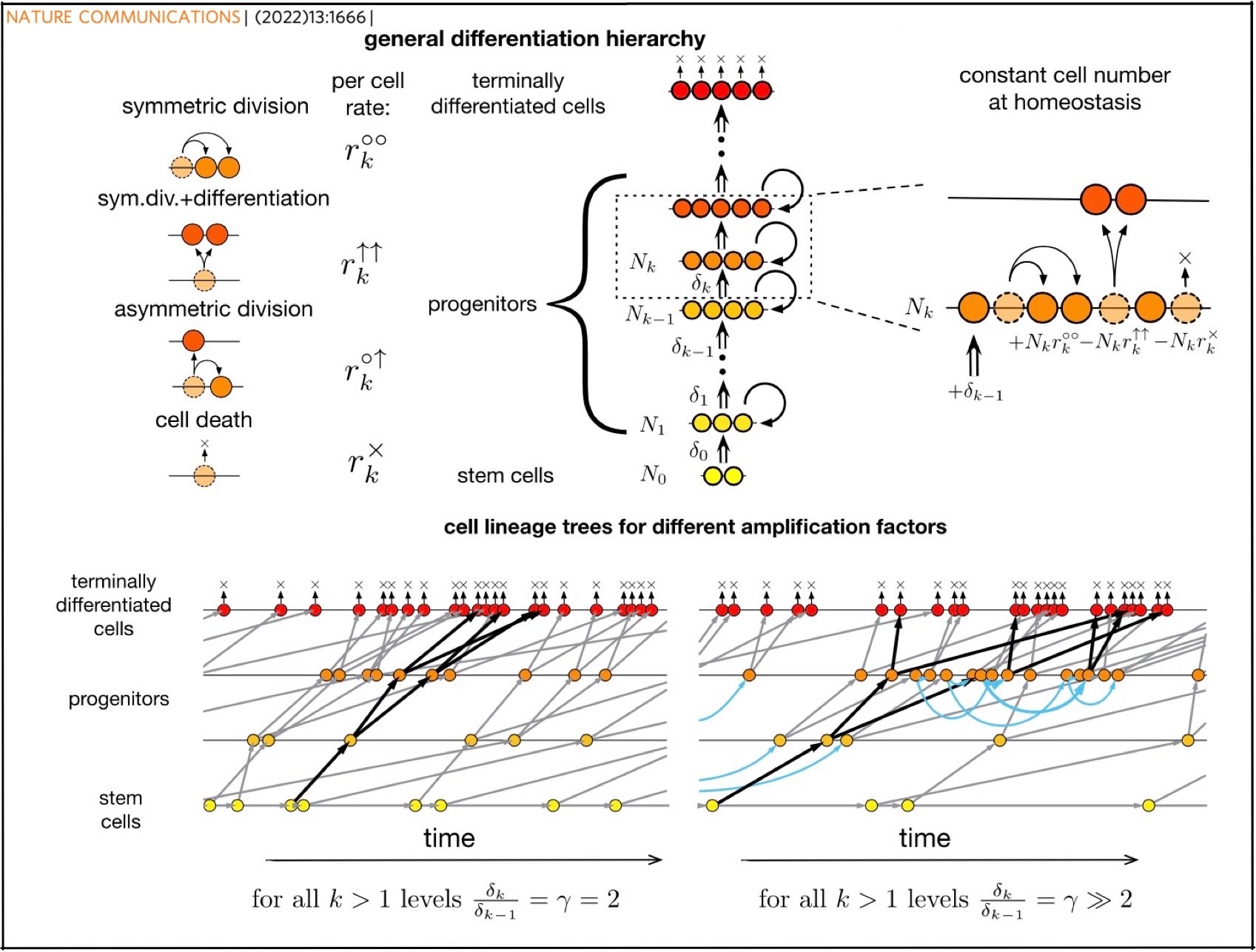
2021:
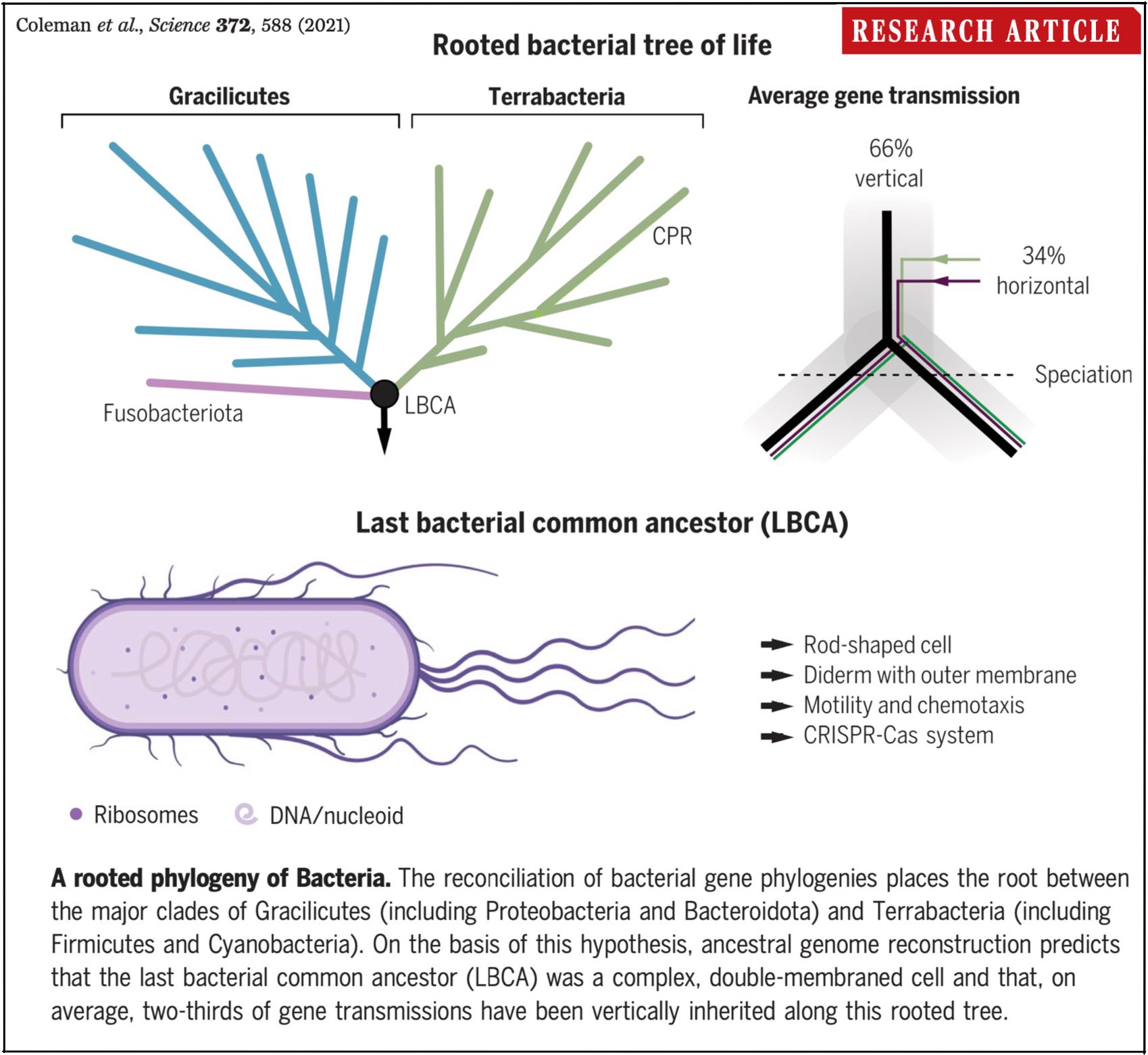
Our new paper "A rooted phylogeny resolves early bacterial evolution" is out in Science!
2020: We are looking for postdocs to work on the origin of eukaryotes in a project funded jointly by the Simons and Moore Foundations, please click (Moore postdoc) for deatils! We are looking for postdocs to work on the ERC "GENECLOCKS" project! Please click (postdocs) for deatils Two new papers out in MBE! Schrempf, Lartillott, Szöllősi: "Scalable empirical mixture models that account for across-site compositional heterogeneity Morel, Kuzlov, Stamatakis, Szöllősi: "GeneRax: A Tool for Species-Tree-Aware Maximum Likelihood-Based Gene Family Tree Inference under Gene Duplication, Transfer, and Loss " Our new paper "Mutants go extinct under a threshold proliferation advantage." is out in PNAS. More on somatic evolution coming soon! 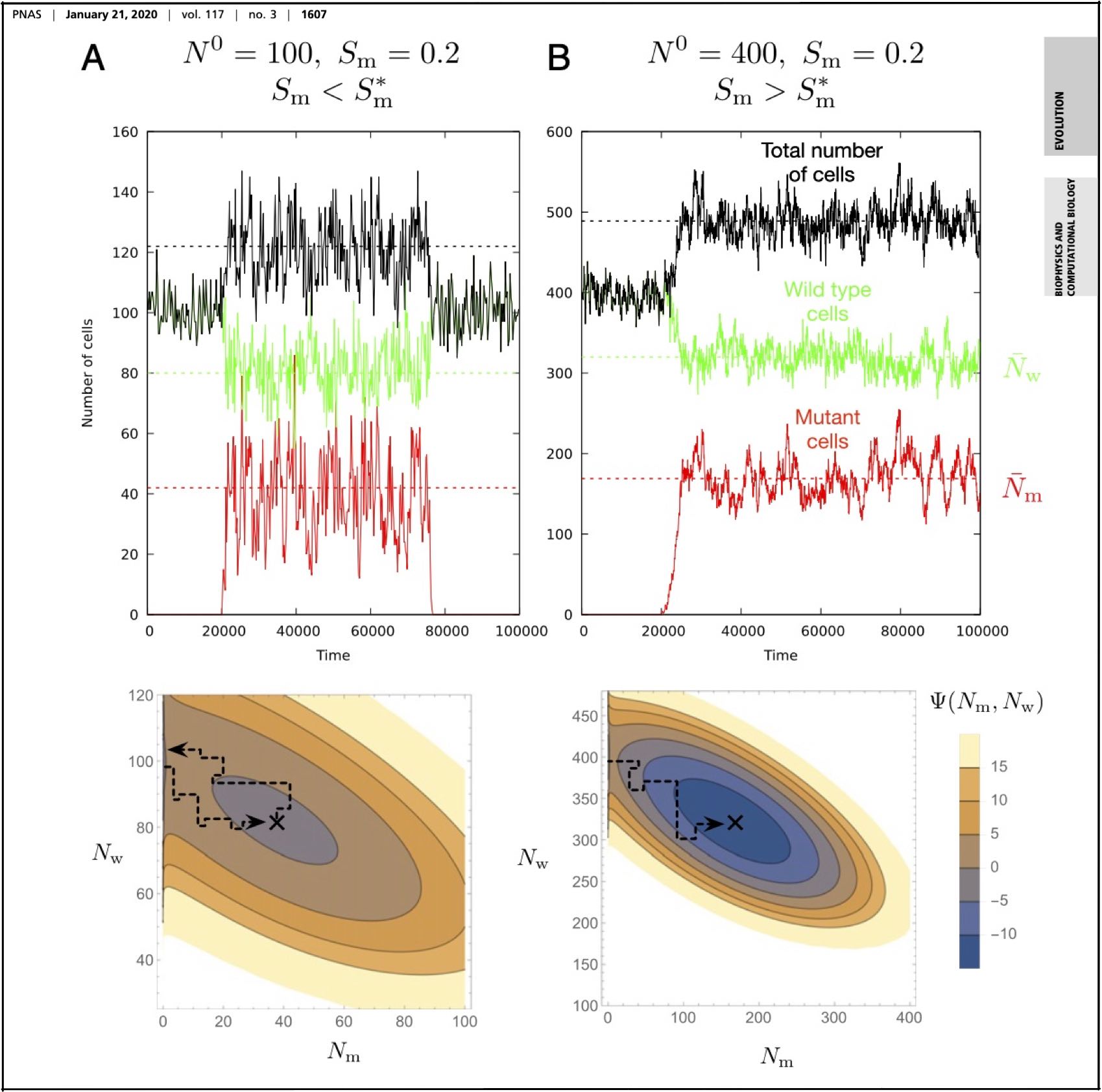
2019:
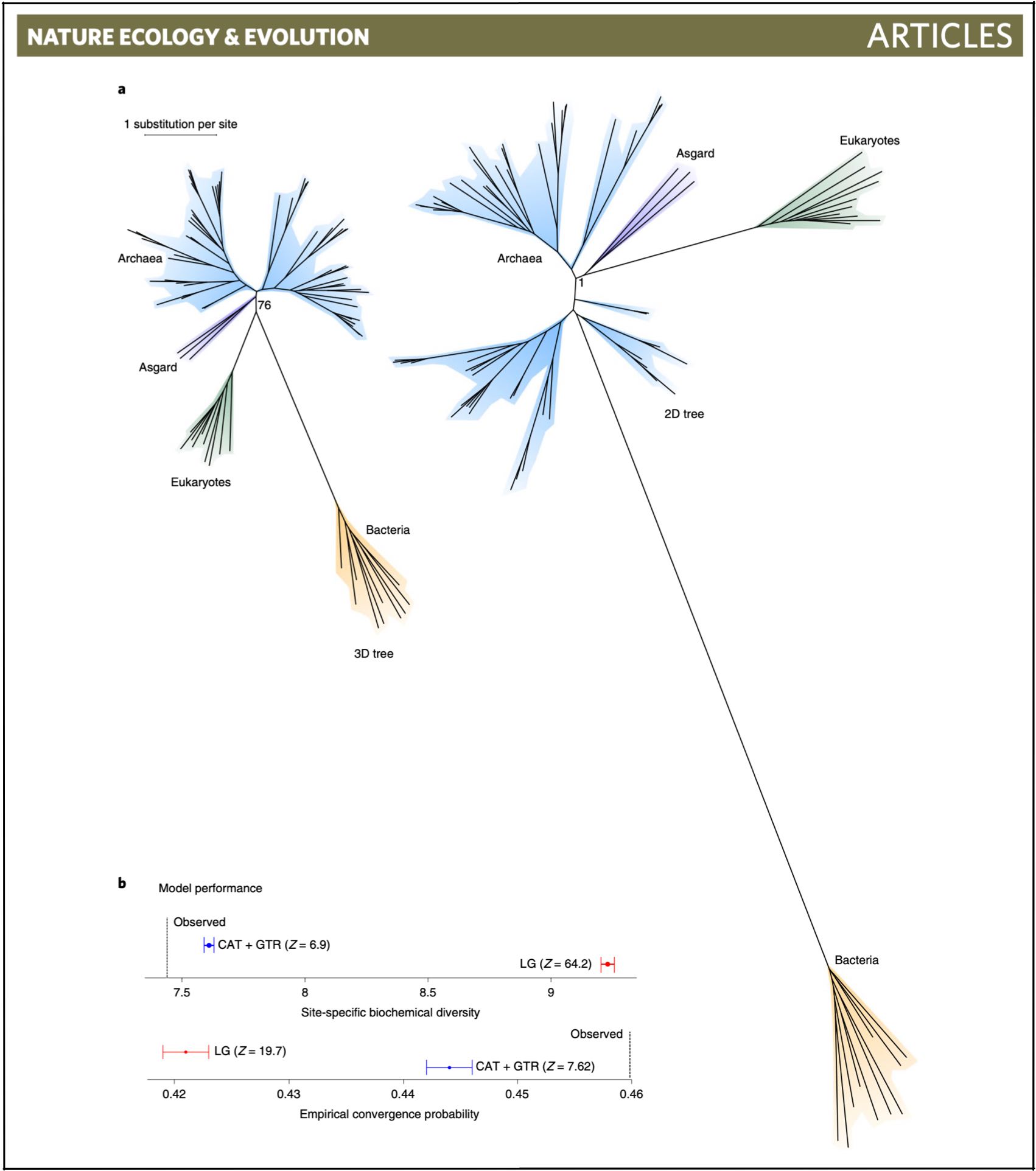
Our new paper "Phylogenomics
provides robust support for a two-domains tree of life" (pdf)
is out in Natrue Ecology and Evolution.
We have two exciting new phylogenetic methods out on github (and BioRχiv)! "GeneRax" developed with Benoit Morel and the guys at "Exelixis lab" allows you to infer species tree aware gene trees straigth from the alignment and "EDCluster" a new, scalable method for finding empirical distribution mixture models (which can be used with IQ-TREE, Phylobayes, and RevBayes and out perform C10-C60), developed with Domink Schrempf and Nicolas Lartillot. 2018: Our paper "Gene transfers can date the tree of life" is out in Nature Ecology and Evolution online and was recently covered in Quanta Magazine: Chronological Clues to Life's Early History Lurk in Gene Transfers. 2017: We are looking for postdocs and PhD students to work on the ERC "GENECLOCKS" project! Please click (postdocs) or (PhDs) for deatils Our new paper "Integrative modeling of gene and genome evolution roots the archaeal tree of life" is out in PNAS! (Open Acces) I am looking for a postdoc to work on Physics of Cancer! Please click here for details. Our new paper "How long does Wolbachia remain onboard?" was recently published in MBE. (link to pdf) Our new paper "Hierarchical tissue organization as a general mechanism to limit the accumulation of somatic mutations" is to apper in Nature Communications. (Open Acces) 2016: Our review "Horizontal Gene Transfer and the History of Life" is to appear in Cold Spring Harbor Perspectives in Biology. (link to pdf) Our paper describing extensive Lateral Gene Transfer among Fungi in the Philisophical Transactions special issue "Eukaryotic origins: progress and challenges" is now online. (Open Acces) and before .. I had the opportunity to give the first in a series of three talks in phyloseminar.org's Mini-course on genome-scale phylogeny. |
| Gergely J. Szöllõsi | ||
 |
|
|
 ORCiD ID:
ORCiD ID:  Google Scholar:
Google Scholar: